Part 4: Towards a metrology of aging based on telomeres.

Measuring aging: Telomeres of physiological age witnesses
Measuring telomere length is a very accurate tool for determining the physiological age of our cells and organs.
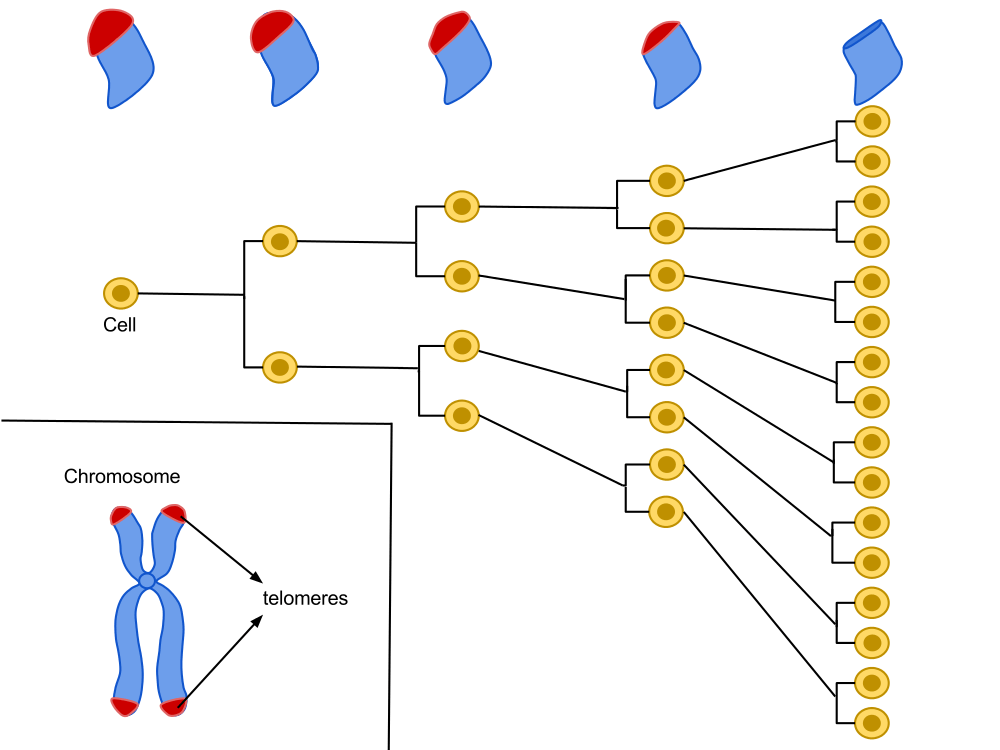
Located at the ends of our chromosomes, telomeres are highly repetitive sequences comprising the sequence of TTAGGG nucleotides. A cell is programmed to divide a certain number of times and, with each cell division, these chromosome ends shorten a little more, until they reach a critical size. The cell will then stop its growth to enter senescence.
Lifestyle, eating habits, stress, environmental factors as well as age can accelerate the shortening of telomeres. Measuring their length can therefore provide valuable information on the progress of the real age of our cells, as well as on the factors influencing it. For all these reasons, measuring telomeres is a remarkable metrological means of the degree of aging of an organism, and it can encourage individuals to take measures to preserve their health.
Hayflick's work has shown that there is a division capital for several cell lineages. It is proportional to the longevity of the species and varies between individuals of the same species. With each cycle of cell division, the end of the chromosomes (telomere) loses a DNA fragment. After several divisions, telomere function, which helps maintain DNA stability, is impaired, which could be the bedrock of the “biological clock.”
The sample to be tested : the measurement of telomere length can be done on cellular or tissue samples, however we will favor carrying out this measurement on the telomeres of leukocytes, a type of cells present in the blood, allowing minimally invasive sampling.
Five main methods of measuring telomeres for aging metrology
Southern blots of Terminal Restriction Fragments (TRFs)
Developed in 1990, it is the most archaic technique, but also the most widely used. This technique estimates the average number of terminal TTAGGG repeats carried by all chromosomes. It is based on the hybridization technique in situ which identifies the TTAGGG repeats capping telomeres. Telomeric DNA is digested by restriction enzymes, resulting in several small DNA fragments of different sizes. In order to know the size of each telomeric DNA fragment, Southern Blot is used.
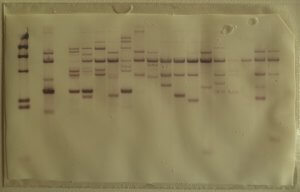
To do this, the telomeric fragments obtained after digestion are separated by gel electrophoresis then transferred to a nitrocellulose or nylon membrane, solid supports which facilitate the denaturation of the DNA (change from double-stranded to single-stranded form). and its hybridization with a probe. This step is crucial in order to allow the hybridization of a radioactive probe (DNA fragment complementary to the desired sequence). The position of the restriction fragments is then revealed by radiography and their size is estimated by comparison between the distance they have traveled in the gel and that covered by fragments of known length [1][2].
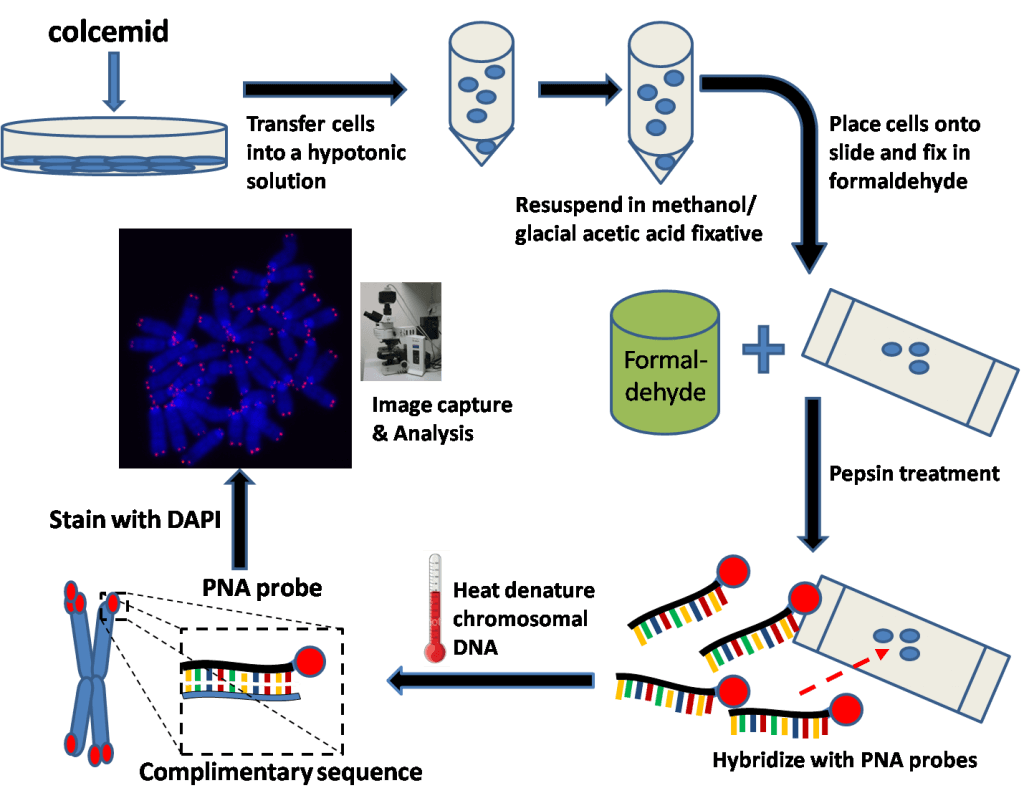
Q-Fish
It is also a hybridization in situ , but unlike Southern Blot, the probe is not radioactive, but fluorescent. The disadvantage of this method is that it requires a large quantity of DNA, up to 20 μg, whereas a few ng are sufficient for other techniques such as Q-PCR [3][4].
Flow-FISH
The Flow-FISH technique is similar to Q-FISH except that it allows direct analysis of telomeres without first DNA extraction. Subsequently, the labeled cells are sorted in a flow cytometer. This method is very expensive and rarely used, because it requires significant equipment. Nevertheless, it remains very interesting: it makes it possible to reduce the purification steps and allows the simultaneous analysis of telomere length in different cell types [7][8].
TAT for measuring telomere length
TAT® assesses telomere length individually. TAT® technology is based on fluorescent hybridization in situ at high throughput, enabled by a High Content Screening (HCS) approach. TAT® provides a comprehensive measurement of telomere length via a histogram. It gives information on telomeres from the shortest to the longest, gives the frequency by length class, the average as well as the median values [9].
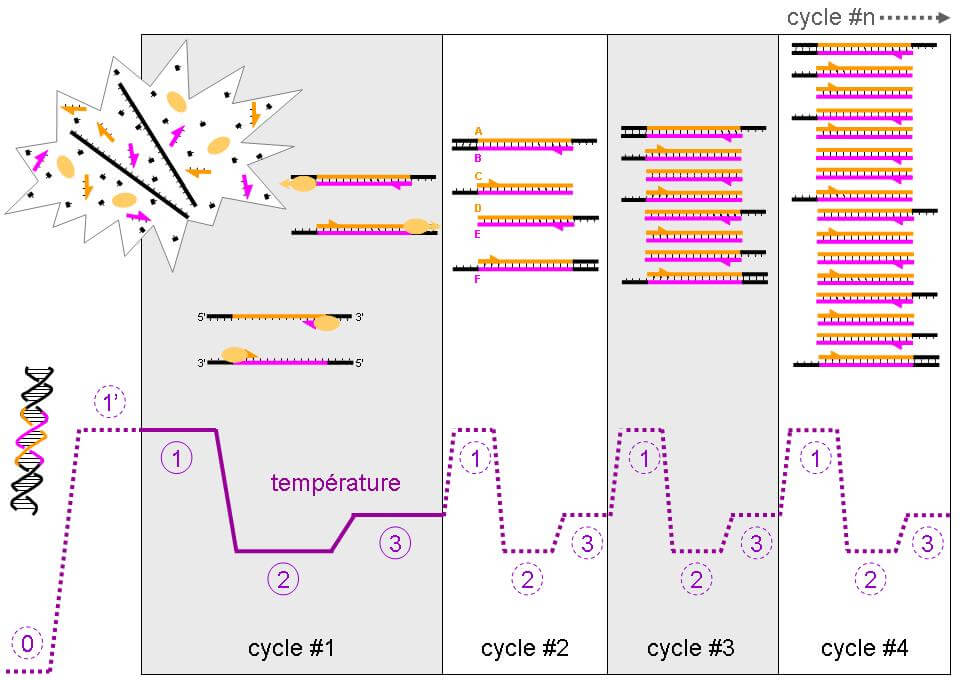
PCR-Q
Developed in 2003, PCR-Q is a faster and easier technique. This technique is based on the principle of hybridization. A DNA sequence is extracted then placed in a machine which will allow three different steps to be carried out (repeated around forty times): denaturation (passage of double-stranded DNA to single-stranded), hybridization with the probe then an amplification of the hybridized product. The probe is a DNA fragment complementary to the sequence of interest and will make it possible to amplify, that is to say increase the number of copies, of this gene. The faster a DNA sequence is amplified, the more it is present in a given sample.
Thanks to several parameters, in particular the ratio of the number of copies of the telomeric motif (T) relative to a control gene (S) (T/S), we can determine the number of TTAGGG repeats, characteristic of telomeric DNA [2 ][5][6].
STELA ( Single TELomere Length Analysis )
This technique differs from Q-PCR in that it makes it possible to amplify, not a telomere fragment but its entire sequence. Its principle is the same for the rest of the procedure. However, it is longer because it requires analysis chromosome by chromosome and not on total DNA [2][7][8].
Telomere length measurement and aging metrology
PCR techniques seem to be the best in terms of reproducibility, but like methods based on hybridization in situ , they cannot amplify telomeres of more than 25 kb. It is possible to observe telomeres of more than 50 kb, but in this case we prefer the Southern Blot.
We know that not all organs of the body age in the same way, and even if leukocytes are a representative cell type for the study of aging, the tests opposite essentially analyze cell populations coming from a blood sample, and that still remains too reductive. Analysis of telomeric length based on a tissue sample (biopsy) is possible, but requires medical supervision and a very invasive procedure.
Where can we request an analysis of our telomeres?
At Medfuture, we have cutting-edge, proprietary technology that measures more than 100,000 individual telomeres. More information
SOURCES:
Katidja Allaoui on http://www.longlonglife.org/
Sources:
[1] Chatterjee, S. (2017). Telomeres in health and disease. Journal of oral and maxillofacial pathology: JOMFP, 21(1), 87.
[2] Blasco, M.A. (2007). Telomere length, stem cells and aging. Nature chemical biology, 3(10), 640-649.
[3] Leri, A., Franco, S., Zacheo, A., Barlucchi, L., Chimenti, S., Limana, F., … & Blasco, MA (2003). Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. The EMBO journal, 22(1), 131-139.
[4] Cawthon, RM, Smith, KR, O'Brien, E., Sivatchenko, A., & Kerber, RA (2003). Association between telomere length in blood and mortality in people aged 60 years or older. The Lancet, 361(9355), 393-395.
[5] Canela, A., Vera, E., Klatt, P. & Blasco, MA High-thoughput telomere length quantification by FISH and its application to human population studies. Proc. Natl. Acad. Sci. USA 104, 5300–5305 (2007).
[6] Teyssier, JR, Ragot, S., Donzel, A., & Chauvet-Gelinier, JC (2010). Telomere length in the cortex of patients with depressive disorders. L’Encephale, 36(6), 491-494.


